Unsupervised Learning
@fig:unsupervised-learning
Unsupervised learning is a type of machine learning that involves training an algorithm on a dataset that contains inputs or features, but no corresponding outputs or labels. The algorithm is left to find patterns, relationships, and structure in the data on its own, without any guidance or supervision from a labeled dataset.
One of the main goals of unsupervised learning is to discover hidden patterns or features in the data that can be used for further analysis or modeling. This can include identifying clusters or groups of similar data points, reducing the dimensionality of the data, or detecting anomalies or outliers.
One of the key concepts in unsupervised learning is the idea of representation learning, which refers to the process of finding a compact and meaningful representation of the data that captures its underlying structure. This can be achieved through techniques such as dimensionality reduction, feature extraction, and feature learning.
Unsupervised learning algorithms can be broadly categorized into two types: clustering and dimensionality reduction. Clustering is used for grouping similar instances together, while dimensionality reduction is used for reducing the number of features used to represent the data while still keeping the most important information.
Another important concept in unsupervised learning is the idea of self-supervised learning, it's a type of unsupervised learning that uses the structure of the data itself as the supervision signal, rather than external labels.
Unsupervised learning is widely used in various applications, such as natural language processing, computer vision, and anomaly detection. It can be a useful tool for exploring and understanding complex datasets, and it often serves as a preprocessing step for supervised learning tasks.
Unsupervised Machine Learning Process
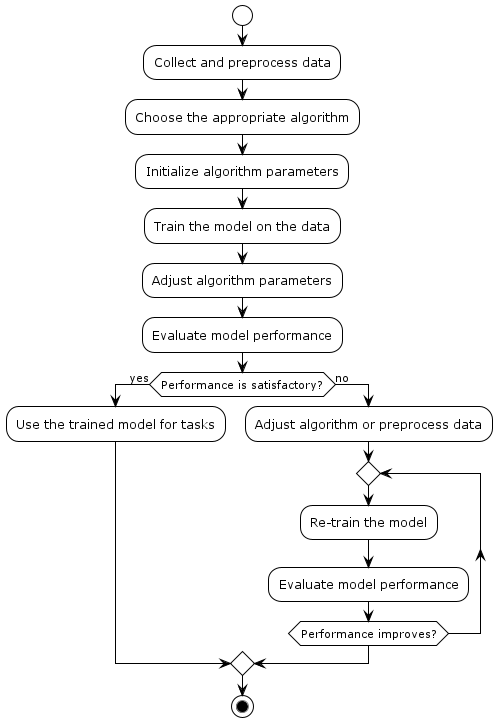
unsupervised-machine-learning-process represents the process of unsupervised machine learning in a simple and visual way. Here's an explanation of each step:
-
Collect and Preprocess Data: This is the initial step where you gather the raw data that will be used for training your unsupervised learning model. Data preprocessing involves cleaning, transforming, and preparing the data for analysis.
-
Choose the Appropriate Algorithm: Depending on the problem you're trying to solve, you select a suitable unsupervised learning algorithm. Common algorithms include K-means clustering, hierarchical clustering, and dimensionality reduction methods like PCA (Principal Component Analysis).
-
Initialize Algorithm Parameters: Set initial values for any parameters that the chosen algorithm requires. These parameters may affect the behavior of the algorithm and its convergence.
-
Train the Model on the Data: Apply the chosen unsupervised learning algorithm to the preprocessed data. The algorithm will attempt to find patterns, relationships, or structures within the data without using any labeled examples.
-
Adjust Algorithm Parameters: After the initial training, you might need to fine-tune the algorithm's parameters to achieve better results. This step involves iterative experimentation to optimize the algorithm's performance.
-
Evaluate Model Performance: Measure how well the trained model is capturing the underlying patterns in the data. Common evaluation metrics depend on the specific algorithm and its objectives, such as within-cluster variance for clustering algorithms.
-
Performance Check: Determine if the model's performance is satisfactory based on the evaluation metrics. If the performance meets your requirements, you can move on to using the trained model for tasks.
-
Adjust Algorithm or Preprocess Data: If the performance is not satisfactory, you may need to adjust the algorithm's parameters again or revisit the data preprocessing steps to enhance the quality of the input data.
-
Iterative Re-training: In case performance isn't satisfactory, you enter an iterative loop where you retrain the model with adjusted parameters or preprocessed data. This process continues until the model's performance reaches an acceptable level.
-
Use Trained Model for Tasks: Once the model's performance is satisfactory, you can utilize the trained model to perform various tasks, such as making predictions, segmenting data, or reducing dimensionality for downstream applications.
Remember that the specific steps and details may vary depending on the algorithm and problem domain you're working with.
Learning Methods
Some common types of unsupervised learning are:
- Clustering
- K-Means Clustering
- Hierarchical Clustering
- Density-Based Clustering
- Singular Value Decomposition (SVD)
- Principal Component Analysis (PCA)
- Self-Organizing Maps (SOM)
- Latent Dirichlet Allocation (LDA)
- Association Rule Learning
- Expectation Maximization (EM)
We now look at the details of these learning methods in detail.
Clustering
Clustering is a type of unsupervised learning where the goal is to group similar data points together into clusters. The algorithm works by dividing the data into clusters such that data points within a cluster are as similar as possible to each other, and as dissimilar as possible to data points in other clusters.
In clustering, the algorithm does not have any prior knowledge of the structure of the data or the number of clusters, it must determine these structures from the data itself. This makes clustering a powerful tool for exploring and understanding the structure of complex and high-dimensional data.
In other words, clustering is the assignment of a set of data (observations) into subsets (clusters), so that data in the same cluster are similar in some sense. For example, one can find that most objects in the natural world fall into one of two categories: things that are brown and run away, and things that are green and don't run away. The first group is called animals, and the second, plants. Clustering is the operation of grouping things together.
There are many different algorithms for clustering, including k-means, hierarchical clustering, and density-based clustering. The choice of algorithm depends on the specific requirements and characteristics of the data.
Clustering has many practical applications, including market segmentation, image segmentation, and text categorization. In these applications, clustering can help identify patterns and relationships in the data that would not be evident otherwise.
K-means Clustering
K-means Clustering is a parametric approach that aims to partition \(n\) data objects into \(k\) clusters where each data object is assigned to the cluster with the nearest mean. In other words, it tries to find the centers of natural clusters of the data objects.
To provide a formal description, consider a set of data objects \((x_1, x_2, \dots, x_n)\), where each data object is a real vector. K-means clustering aims to partition the \(n\) data objects into \(k\) clusters $$ C = {C_1, C_2, \dots, C_k} (k \leq n) $$
so that the within-cluster sum of squares is minimum:
where \(\mu_i\) is the mean of data objects in \(C_i\). An algorithm that is often used for k-means clustering has the following steps, which are iterated (counted by \(t\)) until a convergence is reached (i.e. the assignments do not longer change).
Hierarchical Clustering
Hierarchical clustering is a non-parametric approach that aims to build a hierarchy of clusters. Approaches to hierarchical clustering generally fall into two types:
- Agglomerative: a "bottom up" approach, in which one starts by inserting each data object into its own cluster, and pairs of clusters are merged as one moves up the hierarchy.
- Divisive: a "top down" approach, in which one starts by inserting all data objects in one cluster, and splits are performed recursively as one moves down the hierarchy.
In order to determine which clusters should be merged, or where a cluster should be split, a measure of similarity/dissimilarity (e.g. a measure of distance) is needed between sets of data objects. Some commonly used measures of distance for hierarchical clustering are (where \(a\) and \(b\) are two clusters; and \(a_i\) and \(b_i\) are the data objects of the corresponding clusters):
- Euclidean distance: \(\|a-b \|_2 = \sqrt{\sum_i (a_i-b_i)^2}\)
- Squared Euclidean distance: \(\|a-b \|_2^2 = \sum_i (a_i-b_i)^2\)
- Manhattan distance: \(\|a-b \|_1 = \sum_i |a_i-b_i|\)
- Maximum distance: \(\|a-b \|_\infty = \max_i |a_i-b_i|\)
- Mahalanobis distance: \(\sqrt{ (a-b)^T S^{-1} (a-b) }\) where \(S\) is the covariance matrix.
Density-based Clustering
Density-based clustering is a type of unsupervised learning algorithm that groups data points into clusters based on the density of the data. The idea behind this approach is that data points that are part of a cluster are located close together in the feature space, forming a dense region. On the other hand, data points that are not part of a cluster are dispersed and located far apart from each other.
One of the most well-known algorithms for density-based clustering is DBSCAN (Density-Based Spatial Clustering of Applications with Noise). The algorithm works by defining a neighborhood around each data point and then grouping together data points that are located within dense regions.
The mathematical formulation of the DBSCAN algorithm is as follows:
- For each data point, the algorithm calculates the distance to its k nearest neighbors.
- Based on the distances, a density threshold is set, and data points are classified as core points, border points, or noise points. Core points are those that have at least minPts data points within the density threshold. Border points are those that have fewer than minPts data points within the density threshold but are still close to a core point. Noise points are those that are far from any core points and have no other points within the density threshold.
- The algorithm starts with an arbitrary core point and forms a cluster by adding all core points and border points that are reachable from the core point.
- The process is repeated for all unvisited core points, and the resulting clusters are the output of the algorithm.
The two key parameters in the DBSCAN algorithm are minPts, which determines the minimum number of points required to form a dense region, and the density threshold, which determines the distance between points. The choice of these parameters affects the number and shape of the resulting clusters.
In summary, density-based clustering is a powerful technique for grouping data points into clusters based on the density of the data, and can be applied to a wide range of real-world problems.
| Clustering Method | Pros | Cons |
|---|---|---|
| K-Means | Simple to implement and fast | Sensitivity to initial centroids |
| Hierarchical | Can handle any number of clusters and visual representation | Computationally expensive for large datasets |
| Density-Based | Can handle non-spherical clusters and noise | Difficulty in choosing proper hyperparameters and sensitivity to data scaling |
Table: K-Means, Hierarchical Clustering and Density-Based Clustering
The table above compares three discussed clustering methods. It provides a concise summary of the advantages and disadvantages of each clustering method, which can be useful in choosing the most appropriate method for a particular clustering problem.
Singular Value Decomposition (SVD)
Singular Value Decomposition (SVD) is an unsupervised machine learning type that is used for dimensionality reduction and matrix factorization. SVD is a decomposition of a matrix into three matrices, which can be used to represent the data in a lower-dimensional space while preserving the important information. SVD is widely used in natural language processing, computer vision, and data compression, among other applications.
The mathematical foundations of SVD lie in linear algebra and the theory of matrix factorization. SVD factorizes a matrix into three matrices, which can be used to represent the data in a lower- dimensional space while preserving the important information.
The mathematical formulation of SVD can be represented as follows:
Let \(A\) be a \(m \times n\) matrix. SVD factorizes A into three matrices, \(U\), \(Σ\), and \(V^T\):
where \(U\) is a \(m \times m\) orthogonal matrix, \(Σ\) is a \(m \times n\) diagonal matrix with non- negative singular values, and \(V^T\) is a \(n \times n\) orthogonal matrix.
The singular values in \(Σ\) represent the importance of the corresponding singular vectors in \(U\) and \(V^T\) in representing the data. By keeping only the top \(k\) singular values, the data can be represented in a k-dimensional space, which is a lower-dimensional representation of the data that preserves the important information.
In this way, SVD can be used for dimensionality reduction and matrix factorization, which can be useful for various applications, such as data compression, natural language processing, and computer vision.
Principal Component Analysis (PCA)
Principal Component Analysis (PCA) is an unsupervised machine learning technique that is used for dimensionality reduction and feature extraction. PCA is based on the idea of finding a low- dimensional representation of the data that captures the most important information. PCA is widely used in various fields, such as image and signal processing, data visualization, and machine learning.
The mathematical foundations of PCA lie in linear algebra and the theory of eigenvectors and eigenvalues. PCA finds a set of orthogonal axes, called principal components, that capture the most important information in the data. These principal components are found by computing the eigenvectors of the covariance matrix of the data.
The mathematical formulation of PCA can be represented as follows:
Let \(X\) be a \(m \times n\) matrix, where \(m\) is the number of samples and \(n\) is the number of features. PCA finds a set of \(k\) principal components, where \(k < n\), that capture the most important information in the data.
The covariance matrix of \(X\) is computed as:
where \(X^T\) is the transpose of \(X\).
The eigenvectors and eigenvalues of \(Σ\) are computed, and the \(k\) principal components are found as the \(k\) eigenvectors with the largest eigenvalues. These principal components are used to project the data into a k-dimensional space, which is a lower-dimensional representation of the data that captures the most important information.
In this way, PCA can be used for dimensionality reduction and feature extraction, which can be useful for various applications, such as data visualization, image and signal processing, and machine learning.
Self-Organizing Maps (SOM)
Self-Organizing Maps (SOM) is an unsupervised machine learning technique used for dimensionality reduction and data visualization. SOM is a type of artificial neural network that uses a grid of nodes to represent the data in a lower-dimensional space. SOM is commonly used to visualize and cluster high-dimensional data, and it has applications in various fields, such as image processing, speech recognition, and text classification.
The mathematical formulation of SOM can be represented as follows:
SOM consists of a grid of nodes, where each node is associated with a weight vector. The goal of SOM is to map the high-dimensional data to the grid of nodes such that similar data points are mapped to nearby nodes.
Let \(X\) be a \(m \times n\) matrix, where \(m\) is the number of samples and \(n\) is the number of features. Each node in the SOM grid is associated with a weight vector, \(W_i\), where \(i\) is the index of the node. The distance between a data point, \(x_j\), and a weight vector, \(W_i\), is computed as:
where \(||\dots||^2\) is the Euclidean distance.
The best-matching node, \(BMU\), is found for each data point as the node with the smallest distance:
The weight vectors of the BMU and its neighboring nodes are then updated using a learning rule that takes into account the proximity of the nodes to the \(BMU\):
where \(\alpha\) is the learning rate, which decreases over time.
The process is repeated for multiple iterations, and the weight vectors of the nodes converge to a representation of the data in a lower-dimensional space. In this way, SOM can be used for dimensionality reduction and data visualization, which can be useful for various applications, such as image processing, speech recognition, and text classification.
Latent Dirichlet Allocation (LDA)
Latent Dirichlet Allocation (LDA) is a probabilistic generative model typically used for topic modeling in natural language processing. LDA is also used in various applications such as text classification, information retrieval, and sentiment analysis. In topic modeling, it assumes that a document is a mixture of topics, and each topic is a probability distribution over words in the vocabulary. The goal of LDA is to learn these underlying topic distributions that generated a corpus of documents.
The mathematical formulation of LDA involves the following steps:
- Initialize two hyperparameters: alpha (document-topic density) and beta (topic-word density).
- For each document, randomly assign a topic to each word in the document.
- For each topic, randomly assign a word from the vocabulary as its "prototype".
- Repeat until convergence:
- For each word in each document:
* Compute the probability of each topic for the word, based on the current topic
assignments for the document and the current topic-word distributions.
- Sample a new topic for the word, based on the computed probabilities.
- For each topic: * Update the topic-word distribution based on the current topic assignments for all documents and the current word-topic assignments. *Update the document-topic distribution based on the current topic assignments for each document and the hyperparameter alpha.
- For each word in each document:
* Compute the probability of each topic for the word, based on the current topic
assignments for the document and the current topic-word distributions.
The probability of a document, d, being generated from a particular topic, t, is given by the document-topic distribution, theta, which is a probability distribution over all topics:
The probability of a word, \(w\), being generated from a particular topic, \(t\), is given by the topic- word distribution, \(phi\), which is a probability distribution over all words in the vocabulary:
The joint probability of a document, \(d\), and its corresponding words, \(w\), is the product of the document-topic distribution and the topic-word distribution:
The goal of LDA is to maximize the likelihood of the observed corpus of documents, given the topic- word and document-topic distributions:
The optimization is typically done using variational inference or Gibbs sampling.
In summary, LDA is a probabilistic generative model that models each document as a mixture of topics, where each topic is a probability distribution over words in the vocabulary. The model is learned by iteratively updating the topic-word and document-topic distributions, based on the observed corpus of documents.
Association rule learning
Association rule learning is a type of unsupervised machine learning used to identify interesting relationships between variables in large datasets.
Given a set of transactions, association rule learning algorithms aim to find associations between different items that frequently occur together. The two most common metrics used for this purpose are support and confidence.
- Support is defined as the proportion of transactions that contain both items, while
- Confidence is defined as the proportion of transactions containing the first item that also contain the second item.
An association rule can be represented as \(A => B\), where A and B are sets of items, and the arrow represents a conditional relationship. The strength of an association rule is typically measured by its support and confidence values.
The Apriori algorithm is one of the most popular association rule learning algorithms, which uses a bottom-up approach to discover frequent itemsets and generate association rules. It works by progressively building larger itemsets from smaller ones and pruning those that do not meet a minimum support threshold.
Association rule learning has various applications, including market basket analysis, where it is used to identify items that are frequently purchased together, and recommendation systems, where it is used to suggest related products to customers.
Expectation Maximization
Expectation-Maximization (EM) is an unsupervised machine learning technique used for finding the maximum likelihood or maximum a posteriori (MAP) estimate of the parameters of a statistical model in the presence of missing or latent variables. The algorithm is iterative, alternating between two steps: expectation (E-step) and maximization (M-step).
The mathematical formulation of EM can be represented as follows:
Let \(X\) be the observed data, \(Z\) be the latent variables, and \(\theta\) be the parameters of the statistical model. The goal of EM is to find the maximum likelihood or maximum a posteriori estimate of the parameters, \(\theta\), given the observed data, \(X\), and the latent variables, \(Z\).
The E-step computes the expected value of the complete-data log-likelihood with respect to the current estimate of the parameters, \(\theta\).
The M-step maximizes the expected complete-data log-likelihood from the E-step to update the estimate of the parameters, \(\theta\).
The algorithm iterates between the E-step and the M-step until convergence, where the estimate of the parameters, \(\theta\), no longer changes.
EM is widely used in various applications, such as clustering, dimensionality reduction, and missing data imputation, and it can be used with a variety of statistical models, such as Gaussian mixture models, latent Dirichlet allocation, and factor analysis.
| Technique | Application Areas |
|---|---|
| K-Means Clustering | Image segmentation, customer segmentation, anomaly detection, document clustering |
| Hierarchical Clustering | Biology, social network analysis, document clustering, image segmentation |
| Density-Based Clustering | Spatial data analysis, image analysis, anomaly detection, customer segmentation |
| Singular Value Decomposition | Image processing, recommendation systems, natural language processing, data compression |
| Principal Component Analysis | Image processing, data compression, anomaly detection, bioinformatics |
| Self-Organizing Maps | Image analysis, feature extraction, data visualization, pattern recognition |
| Latent Dirichlet Allocation | Topic modeling, text mining, bioinformatics, image analysis |
| Association Rule Learning | Market basket analysis, cross-selling, customer segmentation, healthcare analytics |
| Expectation Maximization | Image segmentation, text mining, bioinformatics, recommendation systems |
Table: Comparing the application areas of unsupervised learning techniques
The above table provides a brief overview of the application areas for each unsupervised learning technique. The techniques differ in their underlying algorithms and therefore excel in different areas.